Bioinformatics Professor Earns NIH Grant

Dr. Xiuxia Du, an Associate Professor of Bioinformatics at UNC Charlotte’s College of Computing and Informatics (CCI), has been awarded a National Institutes of Health (NIH) Cooperative Agreement Award (U01) as the Principal Investigator (PI) to develop metabolomics data analysis and interpretation tools.
Metabolomics is the comprehensive study and analysis of small molecules, commonly known as metabolites, within cells, biofluids, tissues, or the entire organism. It is estimated that we humans contain thousands of metabolites. They not only keep us healthy but are also important indicators of diseases when we are sick. The function of many of these metabolites are currently unknown. An understanding of what they do and how they do it will inform us what preventative measures we can take to stay healthy and enable early diagnosis of diseases.
Mass spectrometers coupled with chromatography is the primary analytical instruments that scientists use to detect metabolites in metabolomics studies. The capabilities of these instruments have far surpassed our ability to make sense of the mass spectrometry data they generate.
Du’s work is in the development of algorithms and software tools for accurately extracting metabolite information from mass spectrometry-based metabolomics data. It is a critical piece of solving one of the grand challenges of modern science: how genotype (DNA) controls phenotype. Roughly speaking, DNA is used to produce RNA, which gives instructions to generate proteins, which control metabolism. We have cheap, accurate, and relatively easy ways to characterize DNA, RNA, and proteins, but interpreting the mass spectra of metabolites is a vexing problem.
“As a result of this funding,” Du says, “researchers will have a more accurate and robust software tool to detect chemicals in their biological samples.” The significance of this on a global scale is difficult to predict, but in devoting more than $1.35M in a single grant to the cause, the NIH believes it will be substantial. The resulting software tools will allow biologists and chemists to better detect good and bad chemicals in biological samples (blood, urine, tissue, etc.) and address an age-old question in the study of these biological samples: what chemicals are in there and how they make us healthy or sick.
Because metabolomics forms the intersection between genetic pre-dispositions and environmental influences – the nature-nurture balance – its study is key to gaining a better understanding of how genes function and using that knowledge in the discovery of new benefits to our health. Dr. Du’s work greatly facilitates this discovery process that will eventually lead to the discovery of genetic markers for the incidence, transmission and severity of previously unknown or misunderstood diseases. Any time researchers are able to study something in our biology that had been previously unknown or misunderstood, we learn more about what keeps us healthy and what makes us sick.
Dr. Du’s software tool is intended to process data from a wide range of analytical instruments. “I will collaborate with three researchers: Dr. Susan Sumner at UNC-Chapel Hill, Dr. Stephen Barnes from University of Alabama at Birmingham, and Dr. Corey Broeckling at Colorado State University,” Du says. “Each of these collaborators directs a metabolomics core facility at his/her institution and will test the software that my group develops. This collaboration makes extensive testing of the software tool possible.”
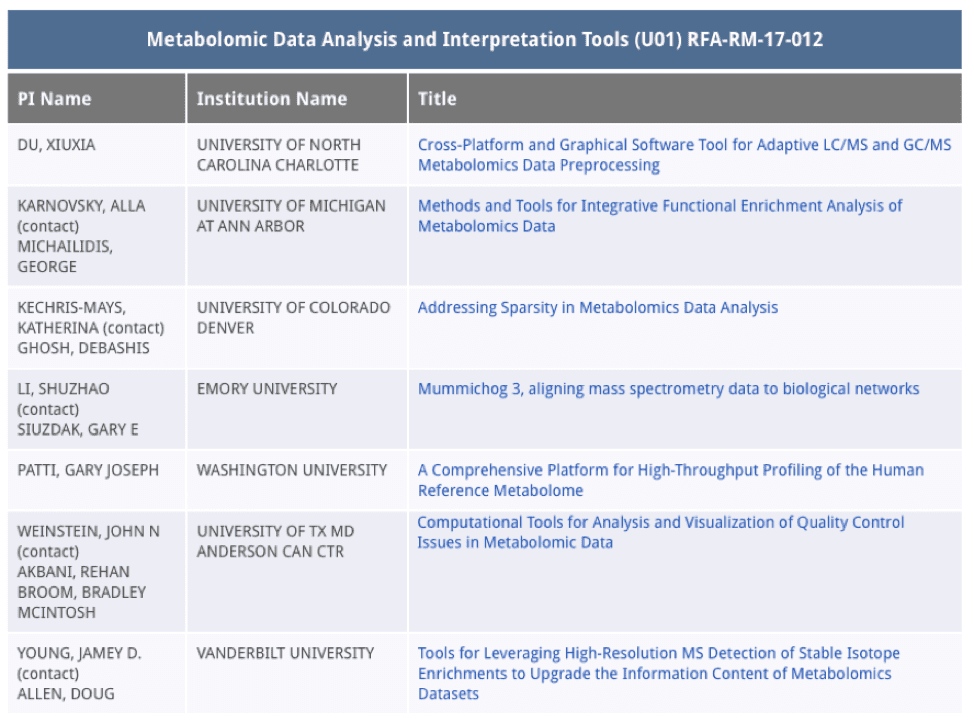
Dr. Du’s award is one of seven awards that the NIH Common Fund Phase II Metabolomics Program has funded for developing Metabolomics Data Analysis and Interpretation Tools. The other six awarded principal investigators are from much better funded and more prestigious programs in the United States (see chart below). As a critical member of this Phase II program, Dr. Du will work across institutional and geographic lines and actively build additional collaborations for the success of the program.
For more News, Notes and Updates, Follow US on Twitter @UNCC_CCI