Larry Mays Seminar Series in Bioinformatics
UNC Charlotte’s Department of Bioinformatics and Genomics (BiG) hosts a weekly seminar series named after Dr. Lawrence Mays (Emeritus Faculty). The Larry Mays Seminars in Bioinformatics covers research within Bioinformatics, Genomics, and Computational Biology. Seminars are free and open to the UNC Charlotte community. The seminars are also a one-credit graduate course (BINF-6600 and BINF-8600 combined). Students enrolled in BINF 6600 or BINF 8600 can see course details on Canvas.
Seminars take place on Fridays at 2:30- 3:30 PM. Seminars are hosted from the Bioinformatics building, room 105. UNC Charlotte’s students, faculty, and staff can CLICK HERE to see more details, inclusing Zoom link information, in our calendar.
Fall 2025 / Spring 2026
Schedule
See the full schedule below or click here to download a PDF version.
Past seminars
Spring 2025
The Larry Mays Seminars in Bioinformatics are hosted by Dr. Denis Jacob Machado this Spring semester of 2025. Please email him at dmachado@charlotte.edu if you have any questions or would like to meet with any of the speakers.
The Spring 2025 edition of the Larry Mays Seminars in Bioinformatics showcases a remarkable diversity of research topics, spanning evolutionary biology, genomics, microbiology, and computational biology. The seminar series features experts investigating a wide range of systems—from Drosophila genetics and microbial ecology to coral symbiosis, venom microbiomes, and RNA biophysics. This breadth of topics highlights the interdisciplinary nature of modern bioinformatics and its applications in understanding complex biological systems.
The speakers represent an impressive array of institutions, including leading research universities such as Duke University, the University of Vermont, and the University of North Carolina, as well as renowned research organizations like the NIH. Additionally, the inclusion of industry-based scientists and researchers from international institutions like the Museum für Naturkunde and the University of Queensland broadens the global perspective of the series.
Beyond the diversity of research topics and institutions, this semester’s lineup also reflects a commitment to representation and inclusivity. The invited speakers come from a range of ethnic backgrounds, genders, and sexual orientations, enriching the seminar series with a variety of perspectives and lived experiences. This diversity not only fosters a more inclusive academic environment but also serves as an important reminder of the value of different voices in shaping the future of bioinformatics and the broader scientific community. By featuring both in-person and virtual talks, the seminar series remains accessible to a wider audience while fostering discussions between students, faculty, and visiting scholars. The diversity of topics, institutions, and speakers ensures a stimulating lineup that captures the rapidly evolving landscape of bioinformatics and its intersection with ecology, evolution, and biomedical sciences.
April 25, 2025: “Integrating sequencing, software, and taxonomy to reveal hidden biodiversity” by Dr. Amrita Srivathsan

Abstract: Despite centuries of exploration, much of Earth’s biodiversity, particularly among insects, remains undocumented. Scalable and affordable methods are urgently needed to discover unknown species at a time of accelerating biodiversity loss. Large-scale DNA barcoding, especially using a reverse workflow where specimens are sequenced first and sorted later, offers a promising solution. Nanopore sequencing enables decentralized and cost-effective barcoding. I present a streamlined workflow using ONTbarcoder, an intuitive software tool that processes MinION data for real-time barcode generation, error correction, and quality filtering. It produces over 10,000 high-quality barcodes per flow cell at under $0.10 each. Flongle supports smaller-scale projects. However, sequencing is only the starting point; barcodes and morphology can be used to delimit species in an integrative framework to facilitate taxonomy. I introduce software for Large-Scale Integrative Taxonomy that visualizes barcode clusters, prioritizes specimens for taxonomic work, and manages taxonomy projects efficiently. This approach connects molecular data to taxonomic practice to accelerate species discovery.
About the speaker: Dr. Amrita Srivathsan is a scientist at the Museum für Naturkunde Berlin’s Center for Integrative Biodiversity Discovery. Her research focuses on enabling species discovery and understanding species interactions. She developed ONTbarcoder, a user-friendly tool for real-time DNA barcoding using nanopore data and has contributed workflows for iDNA-based vertebrate monitoring. More recently, she has developed a platform for Large-Scale Integrative Taxonomy, which connects barcode data to taxonomic workflows and accelerates species delimitation in an integrative framework. Amrita holds a Bachelor’s from the National University of Singapore and a PhD from a joint NUS–Imperial College London program.
April 18, 2025: “Pan-cancer mutational signature analysis of 111,711 targeted sequenced tumors using SATS” by Dr. Bin Zhu (NIH/NCI)

Abstract: Tumor mutational signatures are informative for cancer diagnosis and treatment. However, targeted sequencing, commonly used in clinical settings, lacks specialized analytical tools and a dedicated catalogue of mutational signatures. Here, we introduce SATS, a scalable mutational signature analyzer for targeted sequencing data. SATS leverages tumor mutational burdens to identify and quantify signatures in individual tumors, overcoming the challenges of sparse mutations and variable gene panels. Validations across simulated data, pseudo-targeted sequencing data, and matched whole-genome and targeted sequencing samples show that SATS can accurately detect common mutational signatures and estimate their burdens. Applying SATS to 111,711 tumors from the AACR Project GENIE, we created a pan-cancer mutational signature catalogue specific to targeted sequencing. We further validated signatures in lung, breast and colorectal cancers using an additional 16,774 independent samples. This signature catalogue is a valuable resource for estimating signature burdens in individual targeted sequenced tumors, facilitating the integration of mutational signatures with clinical data.
About the speaker: Dr. Bin Zhu is a Senior Investigator at Division of Cancer Epidemiology and Genetics of National Cancer Institute. His research integrates statistics and genomics to extract mutational signatures and characterize their etiologies across different study designs and platforms.
April 11, 2025: A talk in two parts (I: Bioinformatics Landscape in 2025 and II: Finding the Causes of Cancer Mutations) by Dr. Steve Rozen

Abstract: In part 1 of this talk, Steve Rozen will share his perspectives on the rapidly evolving landscape of bioinformatics in basic and clinical research. One driver of this rapid evolution is deep learning, which offers new approaches to understanding biology while simultaneously changing how we analyze data and develop software. This is coupled with an expanding repertoire of ever-higher-throughput “omics” data-generation technologies that demand new algorithms and software. In part 2, he will talk about his work on cancer genomics and, in particular, computational approaches for investigating the causes of somatic mutations in cancers. These approaches depend on the characteristic patterns of mutations caused by external exposures or internal mutagenic processes. In turn, understanding the causes of mutations in cancer sheds light on cancer epidemiology and tumor evolution.
About the speaker: Dr. Steve Rozen‘s research has spanned bioinformatics, human genetics, and cancer “multi-omics”. His work on bioinformatics and cancer was part of collaborations that led to multiple papers, including 5 in Nature Genetics. This research was recognized by the American Association for Cancer Research 2018 Team Science Award for work on aristolochic acid mutagenesis in liver and urinary tract cancers and for studies of biliary tract cancers, lymphomas, and gastric cancers. Within genomics, he has recently studied mutational signatures as tools for discovering the causes of mutations in cancer and for studying mechanisms of DNA damage and repair. His reference paper on mutational signatures has been cited > 3000 times since 2020. Previously, Rozen also created and maintained the widely-used Primer3 software for PCR primer design and worked extensively on identifying mutations in the human Y chromosome and their clinical consequences.
Mar. 28, 2025: “Intensive transmission in wild, migratory birds drove rapid geographic dissemination and repeated spillovers of H5N1 into agriculture in North America” with Dr. Lambodhar Damodaran

Abstract: Since late 2021, a panzootic of highly pathogenic H5N1 avian influenza virus has driven significant morbidity and mortality in wild birds, domestic poultry, and mammals. To recover how these viruses were introduced and disseminated in North America, we analyzed 1,818 Hemagglutinin (HA) gene sequences sampled from North American wild birds, domestic birds and mammals from November 2021-September 2023 using Bayesian phylodynamic approaches. Using HA, we infer that the North American panzootic was driven by ∼8 independent introductions into North America via the Atlantic and Pacific Flyways, followed by rapid dissemination westward via wild, migratory birds. Transmission was primarily driven by Anseriformes, shorebirds, and Galliformes, while species such as songbirds, raptors, and owls mostly acted as dead- end hosts. Backyard birds were infected ∼10 days earlier on average than birds in commercial poultry production settings, suggesting that they could act as “early warning signals” for transmission upticks in a given area. Our findings support wild birds as an emerging reservoir for HPAI transmission in North America and suggest continuous surveillance of wild Anseriformes and shorebirds as crucial for outbreak inference.
About the speaker: Dr. Lambodhar Damodaran is a postdoctoral research fellow in the lab of Dr. Louise Moncla at the University of Pennsylvania. HIs current research focuses on the epidemiology of RNA viruses with zoonotic potential such as highly pathogenic avian influenza and canine influenza virus. He is particularly interested in poorly sampled pathogens that affect urban and peri-urban animal hosts. He earned his PhD in Bioinformatics at the University of Georgia in 2023 where he studied the evolution and epidemiology of human seasonal influenzas in North America in the lab of Dr. Justin Bahl. His doctoral research was supported by the IDEAS (Infectious Disease Ecology Across Scales) NSF-NRT graduate research fellowship which trained him in ecological modeling methods. He completed his B.S. in Biology at the University of North Carolina at Charlotte and conducted undergraduate research in the lab of Dr. Daniel Janies studying flavivirus evolution and insecticide resistance in malarial vectors.
Mar. 21, 2025: “In search of the Holy Grail in morphological phylogenetics” with Dr. Ambrosio Torres G.

Abstract: The role of morphological data in phylogenetics has been increasingly marginalized in the genomic era, often perceived as unreliable or insufficient in the face of overwhelmingly large molecular datasets. In this talk, I explore two complementary approaches aimed at redefining the place of morphology in modern phylogenetics. First, I present Balanced Resampling (BR), a new method designed to address contribution biases in total-evidence analyses by extending traditional support measures to account for extreme partition disparity. This approach enables a more balanced evaluation of morphological and molecular partitions, allowing for a more nuanced understanding of their reciprocal contributions. Second, I introduce an AI-powered framework for large-scale morphological data integration, which facilitates the incorporation and coding of hundreds or thousands of characters—including complex traits like color and surface patterns. This method leverages quantitative approaches that align with hypotheses of positional homology derived from geometric morphometric analyses. By tackling these challenges from both a methodological and computational perspective, I propose a vision for a future in which morphological data are not merely ancillary but instead play a central and rigorous role in phylogenetic inference.
About the speaker: Dr. Ambrosio Torres G. (from the Center for Integrative Biodiversity Discovery-CIBD) is a researcher in the field of systematics whose work centers on developing and evaluating phylogenetic methods to reconstruct the historical relationships among organisms. His research involves the use of both molecular and morphological datasets, while also integrating modern sources of information such as genomic and morphometric data, to infer the underlying patterns of speciation and diversification. A significant aspect of his work includes the discovery of new taxa through the development of innovative methods for managing and updating taxonomic databases. This is essential to ensure that the vast amounts of data generated from molecular and morphological studies are accurately utilized, classified, and organized. Overall, Dr. Torres’s research seeks to advance our understanding of the systematics of the tree of life, providing valuable insights to support the preservation of Earth’s remarkable biodiversity.
Mar. 14, 2025: “Evolving in bursts: characterizing the genetic and physiological basis of adaptive tracking in the wild” with Dr. Joaquin C. B. Nunez

Abstract: Fluctuations in the strength and direction of natural selection through time are a ubiquitous feature of life on Earth. One evolutionary outcome of such fluctuations is adaptive tracking, wherein populations rapidly adapt from standing genetic variation. In certain circumstances, adaptive tracking can lead to the long-term maintenance of functional polymorphism despite allele frequency change due to selection. While adaptive tracking is likely widespread, its genetic architecture and strength relative to other evolutionary forces, such as drift, remains poorly understood. In this work, we leveraged the seasonal biology of temperate populations of the model system, Drosophila melanogaster, to study the genetic and physiological basis of adaptive tracking. By sequencing orchard populations over multiple years, we uncovered genome-wide footprints of adaptive tracking and identified that the cosmopolitan inversion In(2L)t is a “hot spot” for seasonal adaptive loci in temperate habitats. Additionally, we examined how seasonal selection varies across different life stages and discovered that heat tolerance in D. melanogaster is life-stage specific, with embryonic thermal tolerance during gastrulation serving as a key target for thermal selection. By combining the results of our genome scan with adaptive introgression experiments, we mapped embryonic heat tolerance to a major-effect locus on chromosome 2R. This locus, located in the regulatory region of the endopeptidase-encoding gene SP70, exhibits clinal and seasonal variation that correlates with environmental factors. Validation experiments using lines from the Drosophila Reference Genome Panel (DGRP) as well as data from the Drosophila Evolution over Space and Time dataset (DEST) showed that selection actively maintains variation at this locus in natural populations. Our findings highlight how both seasonal and life-stage-specific selection shape genomic variation, providing deeper insight into the evolutionary dynamics of fluctuating selection.
About the speaker: Prof. Joaquin C.B. Nunez (Assistant Professor of Biology & Principal Investigator) is an evolutionary biologist and population geneticist at the University of Vermont, where he studies the genetic basis of adaptation, particularly how organisms evolve to cope with seasonally changing environments. During his post-doctoral work (2020-2023) at the University of Virginia with Prof. Alan Bergland, Joaquin examined genetic variation in D. melanogaster under seasonal selection, exploring rapid adaptation and the role of chromosomal inversions in tracking seasonal shifts in fitness. For his graduate research (2015-2020) at Brown University with Prof. David M. Rand, he investigated genetic variation in the barnacle Semibalanus balanoides across various intertidal habitats, focusing on loci under selection due to environmental variation. Joaquin’s undergraduate research (2013-2015) at the University of Miami, with Profs. Margie F. Oleksiak and Doug L. Crawford, explored mitochondrial genomics in the marine teleost Fundulus heteroclitus.
Feb. 28, 2025: “Marine symbionts: genome evolution and global change” with Dr. Raúl Augusto González-Pech

Dr. Raúl A. González-Pech has been at the Pennsylvania State University since 2022 as an Eberly Postdoctoral Research Fellow. Dr. González-Pech’s work focuses on the genomics and evolutionary biology of dinoflagellates, particularly the symbiotic algae that sustain coral reef ecosystems. His research has contributed to our understanding of Symbiodinium and other members of the Symbiodiniaceae family, uncovering how their genomes have evolved to support mutualistic interactions with corals. He has co-authored multiple influential studies, including comparative analyses of dinoflagellate genomes, investigations into adaptive evolution, and paleogenomic approaches to understanding coral reef responses to anthropogenic change.
This talk will be online. However, BINF-6600 and BINF-8600 students are asked to join us in person as usual.
Feb. 21, 2025: “Estimating dietary intake from human fecal metagenomes” with Dr. Sean Gibbons

Dr. Sean Gibbons is a researcher specializing in computational and experimental methods to study host-microbe systems, particularly microbial communities and their roles in health and the environment. He earned his PhD in biophysical sciences from the University of Chicago in 2015, where he explored microbial communities as models for ecological theory, followed by postdoctoral work at MIT developing techniques to analyze eco-evolutionary dynamics in the human gut microbiome. He previously studied microbiology and synthetic biology at Uppsala University in Sweden as a Fulbright Graduate Fellow. Since joining the ISB faculty in 2018, supported by a Washington Research Foundation Distinguished Investigator Award, Gibbons has focused on how microbial ecosystems adapt to environmental changes, especially within the human gut over a person’s lifespan. His lab integrates computational and experimental approaches to investigate interactions between microbial ecology, evolution, and function, aiming to develop personalized interventions for improving human health.
Feb. 14, 2025: “Insect phylogenetics and our shared connection” with Dr. Dominic A. Evangelista

Abstract: I will overview my research in insect phylogenetics over the last 5 years. I will discuss three approaches to phylogenetics: minimal data phylogenomics, maximal data phylogenomics, and maximum computation phylogenomics. In each, I attempt to improve the phylogeny of cockroaches by optimizing signal-to-noise ratios, managing noise in super-sparse matrices, or minimizing error through intense computation. I will end by discussing tests of phylogenetic support for big questions in the insect phylogeny, using a massive dataset (1k+ species, 1k+ genes).
About the speaker: Dr. Dominic A. Evangelista is an evolutionary biologist interested in how biodiversity originates through evolutionary processes. Dr. Evangelista’s current research aims to infer phylogenies using genome-scale data, map biological trends over evolutionary histories, and to improve upon phylogenetic methods. Dr. Evangelista’s past research explored palaeoentomology, tropical community ecology, niche evolution, and species delimitation.
Feb. 7, 2025: “Are there microbes in venom, and why characterizing them matters for our fundamental understanding of systems biology” with Dr. Sabah Ul-Hasan

Abstract: Holobiont theory views a given organism and its associated microorganisms as their own universe. Essentially, we are each walking ecosystems. We can consider Earth as an ecosystem in its entirety or scale this down to a single cell and its molecular functions therein, which in essence is systems biology. Focusing specifically on ecosystems through a host-microbe lens, results to date largely revolve around the gut microenvironment. At 50,000 studies and counting, these results have yielded substantial contributions not only to our understanding of microbiology but also evolution and systems as a whole. Even so, we have only begun to scratch the surface of host-microbe systems and our understanding of symbioses. Venom systems have convergently evolved across ~200,000 animal species over hundreds of millions of years and yet venom microbiome studies are < 0.1% of all microbiome studies known. This talk explores the significance of investigating venom microbiomes towards expanding multiomics technologies, new avenues for natural product discovery, and added model systems to game theory.
Click here to see the slide deck for this presentation (last accessed on Feb. 7, 2025).
About the speaker: Dr. Sabah Ul-Hasan (phonetic pronunciation: Sub-buh Ool-Huss-son) is a bioinformatics scientist in biotech. The focus of their talk is rooted in their doctorate studies, where their research resulted in the proposal of venom microbiomics as an emerging field with follow-up findings to be available in a soon-to-be released review paper amongst The Initiative for Venom Associated Microbes and Parasites Consortium (iVAMP). Dr. Ul-Hasan grew up in Salt Lake City as a first generation American to Pakistani and Indian parents. There, they completed BSc (3) degrees in Biology, Chemistry, and Environmental & Sustainability Studies at the University of Utah. Dr. Ul-Hasan went on to complete an MSc in Biochemistry at the University of New Hampshire with an emphasis in Marine Science, a PhD in Quantitative & Systems Biology as a Eugene Cota Robles Fellow at UC Merced, and a joint Postdoctoral Scholar and Bioinformatics Lecturer role at Scripps Research as a NIH NGIMS Fellow. Aside from a laundry list of accolades to garner some form of credibility, Dr. Ul-Hasan is most concerned with being a kinder person than the day before and practicing ego death. “That little word ‘I’, terrible ‘I’ disease, one of the worst ‘I’ diseases that has even known” – George Washington Carver.
Jan. 31, 2025: “The physical code in RNA sequences” with Amy Gladfelter (Duke University)

Abstract: Most amino acids are encoded by multiple codons, making the genetic code degenerate. Synonymous mutations affect protein translation and folding, but their impact on RNA itself is often neglected. We developed a genetic algorithm that introduces synonymous mutations to control the diversity of structures sampled by an mRNA. The behavior of the designed mRNAs reveals a physical code layered in the genetic code. We find that mRNA conformational heterogeneity directs physical properties and functional outputs of RNA-protein complexes and biomolecular condensates. The role of structure and disorder of proteins in biomolecular condensates is well appreciated, but we find that RNA conformational heterogeneity is equally important. This feature of RNA enables both evolution and engineers to build cellular structures with specific material and responsive properties.
About the speaker: Dr. Amy Gladfelter is a quantitative cell biologist interested in fundamental mechanisms of cell organization. She is a Duke Science and Technology Professor in the Cell Biology and Biomedical Engineering Departments at Duke University. Previously, she was a Professor of Biology and Associate chair at the University of North Carolina at Chapel Hill from 2016-2023. In her research program, she uses microscopy and biophysical and genetic approaches to study syncytial cells of fungi and placenta. She examines how these large cells spatially organize the cytoplasm via biomolecular condensates and sense their shape. She trained at Princeton University (AB) with Bonnie Bassler, Duke University (Ph.D.) with Danny Lew, and UniBasel Biozentrum (post-doc) with Peter Philippsen before starting her independent career at Dartmouth in the Biological Sciences department in 2006, where she was until 2016. She has been honored with the 2014 Graduate Mentoring Award from Dartmouth, the 2015 Mid-Career Award for Excellence in Research from the American Society of Cell Biology, the 2020 Graduate School Mentoring Award from UNC, a 2024 mentoring award at Duke, and was a Howard Hughes Medical Institute Faculty Scholar. She is an elected fellow of AAAS, the American Academy of Microbiology, and the American Academy for Arts and Sciences.
Jan. 24, 2025: “Jointly modeling the effects of evolutionary processes on genomic variation” with Dr. Parul Johri (University of North Carolina at Chapel Hill)

Abstract: Population genomic inference currently does not account for the effects of selection across the genome. We propose that an appropriate evolutionary null model, when performing inference, should jointly account for the evolutionary processes known to be in constant operation in natural populations- genetic drift (as modulated by the demographic history of the population), heterogeneity in recombination and mutation rates, and importantly, selection against deleterious mutations. We show how not accounting for such constantly operating processes affects inference in population genetics. We demonstrate a statistical framework for jointly inferring the contribution of the relevant selective and demographic parameters. Our approach represents an appropriate baseline model for inference in population genetics at large, which is necessary to accurately assess the role of adaptive processes in shaping genomic variation. I conclude by discussing our recent efforts in building such evolutionary baseline models for human pathogens to better understand their demographic and selective histories.
About the speaker: Dr. Parul Johri is an Assistant Professor at the University of North Carolina, Chapel Hill, in the Department of Biology and the Department of Genetics, with an affiliation in the Integrative Program for Biological and Genome Sciences. Before that, she was a postdoc at Arizona State University, advised by Jeffrey D. Jensen. She earned her PhD in the Evolution, Ecology, and Behavior program at Indiana University, Bloomington, under the supervision of Michael Lynch. Prior to that, she obtained a Master’s in Biology from the Tata Institute of Fundamental Research in Mumbai, and a Bachelor of Science in Mathematics from Delhi, India.
Jan. 17, 2025: “Population Genomics in Action: The Fantastic Four Superwomen Leading the Charge in Speciation, Polyploidy, and Domestication Research” with Dr. Arun Sethuraman (San Diego State University)

Abstract: Work in the Sethuraman Lab focuses on developing new statistical methods, software, and pipelines for understanding population structure, relatedness, speciation, and estimating evolutionary history from population genomic data. Here I will showcase the work of the Sethuraman Lab’s Fantastic Four Superwomen PhD students – (1) Tamsen Dunn’s polyploidy genome simulator and inference framework called SpecKS, (2) Margaret Wanjiku’s series of statistical tests to estimate ghost introgression into extant genomes, (3) Alexandra McElwee-Adame’s work on the genomics of domestication in hops (Humulus lupulus L.), and (4) Raya Esplin-Stout’s pipeline for simulating and testing thousands of coalescent demographic models for use with fastsimcoal28, called CoalMiner.
About the speaker: Dr. Arun Sethuraman (phonetic pronunciation: Uh-roon Say-thoo-rah-men) is an Associate Professor of Population Genetics and Bioinformatics at San Diego State University. Dr. Sethuraman is an evolutionary computational geneticist who develops new statistical methods, software, and pipelines to understand speciation, genomic ancestry, and human evolution out of Africa. He is originally from India, where he completed his Undergraduate studies in Computer Science prior to obtaining a Ph.D. in Bioinformatics and Computational Biology and Genetics from Iowa State University and an NIH Postdoctoral fellowship at Temple University with Dr. Jody Hey. More recently, he was a Faculty in the Biology Department at Cal State San Marcos. He is a recent recipient of the NSF CAREER and an NIH R15 AREA award and joined the Biology Department as a Faculty and Director of the Biological and Medical Informatics program at San Diego State University in the Fall of 2021. He also currently serves on the Board of Directors of the Genetics Society of America and the Arnold and Mabel Beckman Scholars Program.
Complete schedule for the Spring of 2025
| Date | Speaker’s name | Speaker’s institution | Speaker’s website | Seminar type |
17-Jan-25 | Dr. Arun Sethuraman | Department of Biology, San Diego State University | http://www.sethuramanlab.com | In-person |
24-Jan-25 | Dr. Parul Johri | University of North Carolina, Chapel Hill | https://www.johrilab.org | In-person |
31-Jan-25 | Dr. Amy Gladfelter | Duke University, Cell Biology | https://gladfelterlab.net | In-person |
7-Feb-25 | Dr. Sabah Ul-Hasan | Biotechnology Industry | https://github.com/ivamp-consortium | Virtual |
14-Feb-25 | Dr. Dominic A. Evangelista | University of Illinois at Urbana-Champaign | https://www.roachbrain.com | In-person |
21-Feb-25 | Dr. Sean Gibbons | Institute for Systems Biology; University of Washington | https://gibbons.isbscience.org | In-person |
28-Feb-25 | Dr. Raúl González-Pech | The University of Queensland, Institute for Molecular Bioscience | https://science.psu.edu/bio/people/rag5851 | Virtual |
14-Mar-25 | Dr. Joaquin C. B. Nunez | University of Vermont | https://www.jcbnunez.org | In-person |
21-Mar-25 | Dr. Ambrosio Torres G. | Museum für Naturkunde – Leibniz Institute for Research on Evolution and Biodiversity | https://www.researchgate.net/profile/Ambrosio-Torres-G | Virtual |
28-Mar-25 | Dr. Lambodhar Damodaran | University of Pennsylvania | https://scholar.google.com/citations?user=chQKUhEAAAAJ&hl=en | In-person |
11-Apr-25 | Dr. Steve Rozen | Duke University School of Medicine | https://steverozen.net/ | In-person |
18-Apr-25 | Dr. Bin Zhu | NIH | https://irp.nih.gov/pi/bin-zhu | Virtual |
25-Apr-25 | Dr. Amrita Srivathsan | Museum für Naturkunde, Leibniz-Institut für Evolutions- und Biodiversitätsforschung | https://www.museumfuernaturkunde.berlin/en/museum/today/team/amrita.srivathsan | Virtual |
Fall 2024
The Fall 2024 seminars were organized by Dr. Abigail Leavitt LaBella (alabell3@charlotte.edu).
- August 30th (virtual): Dr. Emile Gluck-Thaler, University of Wisconsin-Madison
- Starships: a new frontier for fungal biology
- September 6th (virtual): Dr. Alex Cope, Rutgers University
- Integrating population genetic models with emerging functional genomic technologies to reveal the rules of proteome evolution
- September 13th (in person): Dr. Danillo Augusto, UNC Charlotte (Department of Biological Sciences)
- Decoding the Immune System: Insights into Human Disease Susceptibility and Population Genetics
- September 20th (virtual): Dr. Jóse Vargas-Muñiz, Virginia Tech
- Can targeting the fungal septin cytoskeleton improve existing antifungal therapies?
- September 27th (in person): Dr. Parul Johri, UNC Chapel Hill
- October 4th (in person): Dr. Lawrence Uricchio, Tufts University
- October 11th (virtual): Dr. Matthew Mead, Ginkgo Bioworks
- October 18th (virtual): Dr. Cullen Roth, Los Alamos National Laboratory
- October 25th (in person): Dr. Antonis Rokas, Vanderbilt University
- November 1st (in person): Dr. Xiaojun Ren, UNC Charlotte (Department of Biological Sciences)
- November 8th (in person): Dr. Liesl Jeffers-Francis, NC A&T
- November 15th (in person): Dr. Donald Fox, Duke University
- November 22nd (in person): Dr. Lauren Dineen & Dr. Colby Ford, UNC Charlotte
Spring 2024
- 1/19 (in person): Dr. Aaron Bivins, Lousiana State University
- 1/26 (remote): Dr. Liliana Dávalos, Stony Brook University
- 2/2 (in person): Dr. Morgan Carter, UNC Charlotte (CIPHER)
- 2/9 (in person): Dr. Yaniv Brandvain, University of Minnesota
- 2/16 (in person): Dr. Wynn Meyer, Lehigh University
- 2/23 (in person): Dr. Philipp Brand, Rockefeller University
- 3/1/24 (in person): Dr. Juan Vivero-Escoto, UNC Charlotte (CIPHER)
- 3/8/24: No seminar, Spring Break
- 3/15/24 (in person): Dr. Alejandro Damian-Serrano, University of Oregon
- 3/22/24 TBD
- 3/29/24: No seminar, Refresh Week
- 4/5/24: Dr. Bob Goldstein, UNC Chapel Hill
- 4/12/24: Dr. Meredith Yeager, National Cancer Institute
- 4/19/24: Dr. Elise Lauterbur, University of Arizona
- 4/26/24: No seminar, Spring Break
Zoom link for in-person attendance is available upon request. Please e-mail Dr. Laurel Yohe (lyohe1@uncc.edu) for the link.
Fall 2023
I. September 8, 2023: UNC Charlotte’s HPC student group will introduce a new training module for our research clusters.
The Charlotte HPC Group gave the 9/8 seminar. This new student organization is focused on advancing high-performance computational skills for users of all levels across campus. They do this through various means, including in-person workshops, peer support, and online Canvas training. This day, they briefly showed the new module and told us about some of the other activities they have on the horizon. This seminar was informative for people who are new to the cluster or don’t even have an account and for more experienced users who want to expand their knowledge and find additional resources. The module is already available to the 49er community. Please use this form to request additional information.
II. September 15, 2023: Dr. Ward Wheeler talks about multi-armed bandits, Thompson sampling, and machine learning in phylogenetic graph search, among other topics.
Dr. Ward Wheeler visited in person on 9/15 coming from the Division of Invertebrate Zoology American Museum of Natural History, New York, NY, USA. In his talk, he talked about something cool called the multi-armed bandit problem in the context of phylogenetic graph searching. He uses a technique called Thompson sampling on a bunch of ‘search-bandits’ to help focus on search strategies that work best. This smart random sampling method turns out to be super effective in creating really good phylogenetic graphs, and it’s quicker than other random search methods. It’s like an unsupervised machine learning strategy that we can use on all sorts of phylogenetic data, even if we don’t know much about them beforehand.
III. September 22, 2023: Dr. Lucas Czeck talks about population genetic statistics for the next generation of pool sequencing.
In recent decades, so-called Evolve-and-Resequence (E&R) experiments have become a popular approach to survey rapid evolution of populations over multiple generations. These experiments allow us to measure shifts in the allele frequencies of a population in response to new or shifting environmental conditions, such as a changing climate.
Pool-sequencing of several individuals at once is a cost-effective and efficient tool to obtain reliable allele frequencies from a population of thousands to hundreds of thousands of individuals, and is often used in E&R experiments. However, specialized tools to efficiently analyze these data that take sampling noise stemming from the pool-sequencing approach into account were lacking. We developed two software tools to overcome statistical and bioinformatic challenges arising in this context.
First, we present grenepipe, a workflow from raw sequencing data of individuals or pooled populations to genotypes (variant calling) and population allele frequencies. The pipeline automates trimming, mapping, variant calling, and quality control, with a selection of popular software tools in each of these steps, and produces variant calls and frequency tables. While generally applicable to individual sample data, it offers specialized steps for pool-sequencing. Our software downloads all dependencies and runs all steps automatically using a single command line call, and parallelizes processing for computer cluster environments, allowing large datasets to be analyzed efficiently.
Second, to enable inferences of evolutionary signatures from frequency data, we created grenedalf, a C++ command line tool to compute population genetic statistics. It computes unbiased statistics of Fst, Pi, Tajima’s D with pool-sequencing data, far outperforming alternative tools, and fixing long-standing issues in the existing tools. Further it offers novel data exploration tools such as windowed allele frequency spectrum visualizations based on the allele frequencies, and built-in data filters and manipulations. Apart from its speed, it offers many convenience options, such as reading all standard file formats.
These tools are designed for scalability and ease-of-use with contemporary file formats, which we showcase using the GrENE-net.org project, a large-scale Evolve-and-Resequence experiment with Arabidopsis thaliana from across the world. We will present some preliminary results form that experiment, showcasing the use of the above tools, as well as demonstrating how E&R experiments can be used to investigate rapid evolution under differing environmental conditions such as caused by climate change.
Recommended publications:
IV. September 29, 2023: Dr. Cathy Moore (Biosafety Officer at UNC Charlotte’s Division of Research) gives a special presentation.
Dr. Cathy Moore provided Biosafety Training for Graduate Students.
Dr. Moore is the University contact for:
- Handling and working with biological agents on campus
- Institutional Biosafety Committee protocol review and approval process
- Risk assessment and risk mitigation for biological agents and materials
- Use of select agents
- Dual Use of Research Concern
- Biosafety training courses and requirements
- Microbiological and Biomedical facilities renovation and design
V. October 6, 2023: Dr. Viviana Monje-Galvan (University at Buffalo) talks about an atomistic view of protein-lipid interactions at the cell membrane interface.
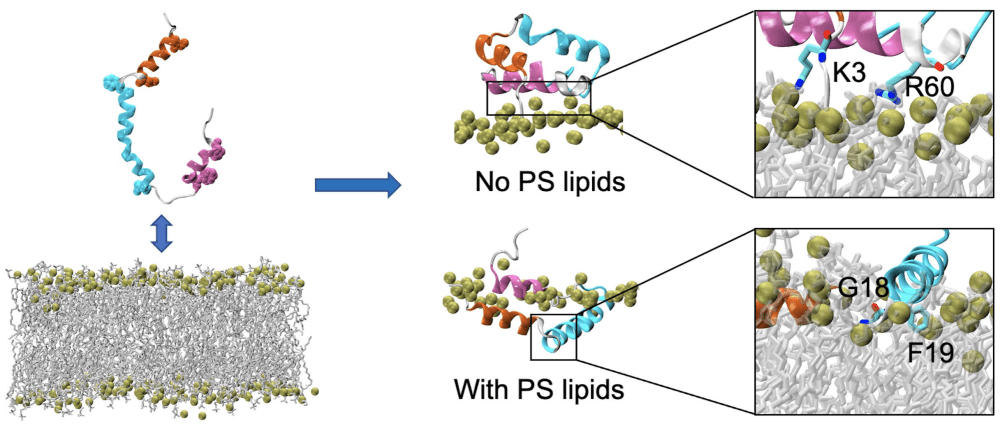
- Talk description: My research group uses all-atom molecular dynamics to study lipid-lipid and lipid-protein interactions. One of our goals is to determine the role of specific lipid species in disease mechanisms. We specialize in developing realistic membrane models that reproduce the physical environment of a given organism or cell organelle using key lipid species for that particular membrane. In this talk, I will present a protein-membrane study in the context of hepatitis C. Specifically, the molecular signature that results upon protein binding and the associated local membrane response when different lipid species are used to model the membrane. These results help us understand the lateral reorganization of lipids around proteins and the potential impacts of chronic hepatitis C and lipid redistribution, particularly near fat storages of the cell.
- Short Bio: Dr. Monje-Galvan is originally from Bolivia; she completed her undergraduate and graduate studies in Chemical Engineering at the University of Maryland-College Park. She completed her postdoctoral training at the University of Chicago, where she continued strengthening her background in computational biophysics. She started her lab at the University at Buffalo in January 2021, where she leads a diverse group of graduate and undergraduate students. She is actively involved with AIChE and the Biophysical Society, as well as her university, in efforts to promote the participation of women and minorities in science and engineering. She is a board member of LatinXinChE, an affinity group within MAC AIChE, AIChE CoMSEF, and is an active volunteer with the Society for Biophysicists of Latinamerica (SoBLA).
VI. October 13, 2023: Dr. Muhammad Iqbal Qureshi (Public Health Reference Laboratory and Khyber Medical University) gives a talk entitled “Genetic Recombination Potentially Drives Astrovirus Diversity in Camelids.”
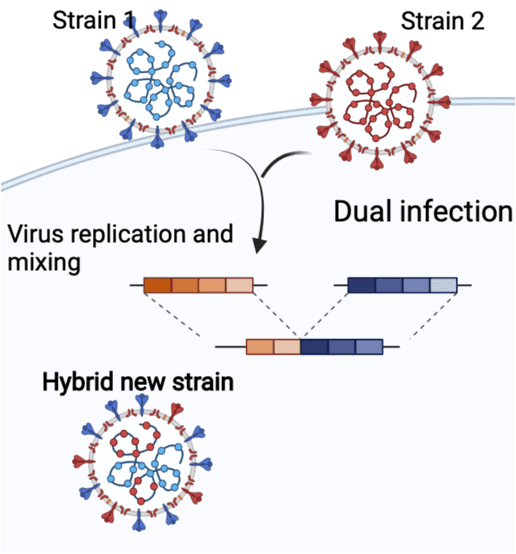
Virus emergence may occur through interspecies transmission and recombination of viruses coinfecting a host, with the potential to pair novel and adaptive gene combinations. Camels harbor diverse ribonucleic acid viruses with zoonotic and epizootic potential. Astroviruses are particularly interesting due to their cross-species transmission potential and endemicity in diverse host species, including humans. We conducted a molecular epidemiological survey of astroviruses in dromedaries from Saudi Arabia and Bactrian camels from Inner Mongolia, China. Our reported sequences expand the known diversity of camel astroviruses, highlighting potential recombination events among the astroviruses of camelids and other host species. Phylogenomic approaches are needed to investigate complex recombination patterns among the astroviruses and infer their evolutionary history across diverse host species.
VII. October 20, 2023: Dr. Brian Cleary (Assistant Professor at Boston University’s College of Engineering) discusses new technologies and algorithmic experimental designs to study intracellular and intercellular circuits.

Dr. Brian Cleary, an accomplished Assistant Professor at Boston University, is a distinguished expert in Cellular Pathways, Tissue Biology, Computational and Systems Biology, Machine Learning, Genomics, and Molecular and Cellular Biology. His research endeavors push the boundaries of algorithmic learning and biological experimentation, aiming to discern the foundational principles governing molecular, cellular, and tissue processes.At the core of his work lies an innovative vision: to explore cellular pathways and tissue biology at scales previously deemed unattainable. This is made possible through a judicious fusion of biological structure and advanced algorithmic and experimental designs. Dr. Cleary’s recent contributions have significantly advanced techniques like Imaging Spatial Transcriptomics and Perturb-seq, offering more streamlined and cost-effective avenues for functional genomics research. His endeavors not only uncover novel regulatory circuits and genetic interactions but also illuminate uncharted realms in diseases such as coronary artery disease, providing profound insights for therapeutic discovery and fundamental biology. In this talk, Dr. Cleary shares invaluable insights and pioneering approaches to address intricate cellular and intercellular processes.
View Dr. Cleary’s complete list of publications chronologically by year.
VIII. November 3, 2023: Dr. Carla Osthoff Ferreira de Barros (National Laboratory of Scientific Computing, Brazilian Ministry of Science, Technology and Innovation) gives a seminar on developing efficient scientific gateways for supercomputer environments supported by deep learning models.
Dr. Carla Osthoff Ferreira de Barros graduated in Electrical Engineering from the Pontifical Catholic University of Rio de Janeiro with a master’s and a doctorate in Computer Engineering Systems from the Federal University of Rio de Janeiro. She is currently a researcher in the area of High-Performance Computing at the National Laboratory for Scientific Computing (LNCC), a professor at the Multidisciplinary Graduate Program at LNCC, a member of the Consultative Committee of the Supercomputer Santos Dumont, coordinates the LNCC National Center for High-Performance Processing (CENAPAD), and the High-Performance Processing Sector of LNCC, which has several collaborative projects in the area of High-Performance Computing. Her research focuses on High-Performance Computing, Parallel Programming, and scientific application performance optimizations.
IX. November 17, 2023: Dr. Grace Wyngaard talks about chromatin diminution and genome stability in a talk that unravels the role of transposable elements in a remarkable adaptation in copepods
Dr. Grace Wyngaard has been working at the “fringes” of science – chromatin diminution in copepods, spending most of her career at James Madison University. This allowed her to pursue what she was most interested in… and only what she was most interested in. It allowed her to take big risks in testing and developing novel hypotheses because she did not have to keep a large lab machine operating each year. In her talk, Dr. Wyngaard will share with us the results of a career in science in which she could work slowly and steed to produce astonishing outcomes.

The publication most relevant to her talk and a broader audience is Drotos et al. (2022), which made the cover of Trends in Genetics. This article covers programmed DNA elimination in ciliates, nematodes, copepods, Japanese hagfish, and sea lamprey. However, programmed DNA elimination (PDE) in copepods was the motivation for preparing this review article.
Dr. Wyngaard’s lab is interested in the following research topics:
- Role of transposable elements in genome reorganization of copepods
- Molecular systematics and cryptic speciation in Cyclops (Crustacea: Copepoda)
- Molecular systematics of marine and freshwater cyclopoid copepods
- Quantitative behavior of DNA during development in Mesocyclops edax (Crustacea: Copepoda)
- Evolution of genome size
X. December 1, 2023: Dr. Joshua David Campbell, from Boston University, will give a talk about his work using computational biology and bioinformatics with applications ranging from identifying early drivers of lung cancer to developing therapeutics for Chronic Obstructive Pulmonary Disease (COPD)

Dr. Joshua D. Campbell is an Associate Professor at Boston University (Division of Computational Biomedicine, Department of Medicine). He is also a member of the BU-BMC Cancer Center and an affiliate member of the Broad Institute of MIT and Harvard. He has received the Boston University Ralph Edwards Career Development Professorship and LUNGevity Career Development Award. He is also a Boston University Clinical and Translational Science Institute (CTSI) KL2 Scholar.
Dr. Campbell’s research interests include computational biology and bioinformatics applied to identifying early drivers of lung cancer. He is also interested in the therapeutic development and pathogenesis of Chronic Obstructive Pulmonary Disease (COPD), the 3rd leading cause of death globally (updated in December 2023).